RecNewPath - DNA recombination, repair and maintenance of genome integrity: new pathways
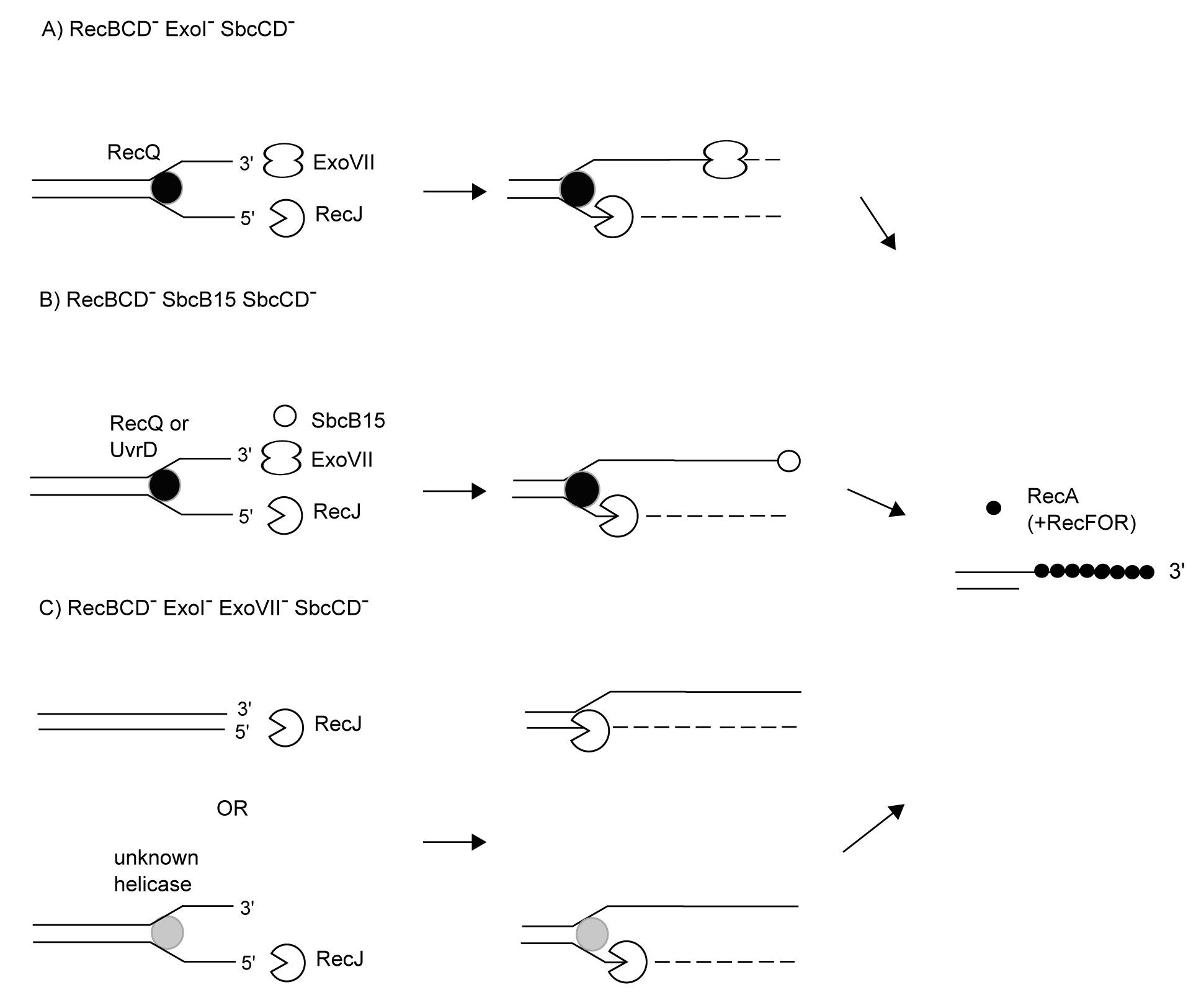
Initiation of recombination at double-strand DNA ends in different variants of the RecF pathway: the role of multiple nucleases and helicases.
Principal investigator
Homologous recombination is an essential biological process that is involved in DNA repair and in the maintenance of genome integrity. RecA protein is a key component of recombinational systems in bacteria. Its homologs are also present in higher organisms including humans. RecA functions in the form of nucleoprotein filaments that are assembled on single-stranded DNA, and which catalyze pairing and strand exchange between two homologous DNA duplexes. In Escherichia coli, the RecBCD and RecFOR protein complexes mediate RecA filament assembly, and consequently, play an important role in recombination. We have recently discovered that some E. coli mutants recombine quite efficiently in the absence of both RecBCD and RecFOR complexes. This recombination is RecA-dependent suggesting the existence of an alternative RecA loading activity. The aim of the project is to genetically characterize this RecBCD- RecFOR-independent (RecBFI) recombination pathway, and gain insight into the novel mechanism of RecA filament assembly.
Although RecA is crucial for efficient recombination in bacteria, the recA mutants of radiation-resistant bacterium Deinococcus radiodurans display a significant residual ability to repair double-strand DNA breaks. Recent results from our group have shown that this RecA-independent repair is quite inaccurate leading to gross chromosome rearrangements. Our project is to identify the key genes/proteins involved in the RecA-independent repair, and to characterize, at the sequence level, gross genome rearrangements in D. radiodurans.
Given that recombination is a fundamental process largely conserved during evolution, our research on bacteria may reveal molecular mechanisms that are applicable to eukaryotic recombination systems. Thus, the results of our project could be instructive for research on cancer and other human hereditary diseases related to defects in DNA recombination functions.
Publications
Zahradka K, Repar J, Đermić D, Zahradka D. Chromosome Segregation and Cell Division Defects in Escherichia coli Recombination Mutants Exposed to Different DNA-Damaging Treatments // Microorganisms, 11 (2023), 3; 11030701, 30. doi: 10.3390/microorganisms11030701
Zahradka K, Repar J, Đermić D, Zahradka D. Genetic analysis of transductional recombination in Escherichia coli reveals differences in the postsynaptic stages of RecBCD and RecFOR pathways // Periodicum Biologorum, 124 (2022), 3-4; 97-106. doi: 10.18054/pb.v124i3-4.23604
Repar J, Zahradka D, Sović I, Zahradka K. Characterization of gross genome rearrangements in Deinococcus radiodurans recA mutants // Scientific Reports, 11 (2021), 1; 10939, 11. doi: 10.1038/s41598-021-89173-9
Buljubašić M, Hlevnjak A, Repar J, Đermić D, Filić V, Weber I, Zahradka K, Zahradka D. RecBCD- RecFOR-independent pathway of homologous recombination in Escherichia coli // DNA Repair, 83 (2019), 102670, 16. doi: 10.1016/j.dnarep.2019.102670
Feliciello I, Zahradka D, Zahradka K, Ivanković S, Puc N, Đermić D. RecF, UvrD, RecX and RecN proteins suppress DNA degradation at DNA double-strand breaks in Escherichia coli // Biochimie, 148 (2018), 116-126. doi: 10.1016/j.biochi.2018.03.005
Đermić E, Zahradka D, Vujaklija D, Ivanković S, Đermić D. 3'-terminated Overhangs Regulate DNA Double-Strand Break Processing in Escherichia coli // G3-Genes Genomes Genetics 7 (2017), 9; 3091-3102. doi: 10.1534/g3.117.043521
Ivanković S, Vujaklija D, Đermić D. Nucleolytic degradation of 3′-ending overhangs is essential for DNA-end resection in RecA-loading deficient recB mutants of Escherichia coli // DNA Repair, 57 (2017), 56-65. doi: 10.1016/j.dnarep.2017.06.024
Repar J, Supek F, Klanjšček T, Warnecke T, Zahradka K, Zahradka D. Elevated rate of genome rearrangements in radiation-resistant bacteria // Genetics, 205 (2017), 4; 1677-1689. doi: 10.1534/genetics.116.196154
Đermić D. Double-strand break repair mechanisms in Escherichia coli: Recent insights // Advances in Genomics and Genetics, 5 (2015), 35-42. doi: 10.2147/AGG.S51699
Diploma Thesis
Hleb A. Genome rearrangements in Deinococcus radiodurans: role of the RecG protein / Hulak N, Zahradka K (mentor); Zagreb, Faculty of Agronomy, University of Zagreb, 2018.
Conferences/Summer Schools
Repar J. DNA double-strand break repair and genome stability in Deinococcus radiodurans // DNA double helix- seventy years later, CePOZiR & HBD meeting Zagreb, Croatia, 28.11.2023. (invited lecture)
Zahradka K, Repar J, Đermić D, Zahradka D. Chromosome segregation and cell division defects in Escherichia coli after induction of double- strand DNA breaks // Power of Microbes in Industry and Environment 2023 : Book of Abstracts / Teparić R, Leboš Pavunc A, Kifer D (ed.). Zagreb: Croatian Microbiological Society, 2023. p. 73-73 (poster)
Zahradka D. RecBCD- RecFOR-independent pathway of homologous recombination in Escherichia coli // FEMS Summer School for Postdocs. Split, Hrvatska 28.08.2019-07.09.2019 (invited lecture)
Buljubašić M, Zahradka K, Hlevnjak A, Repar J, Đermić D, Zahradka D. RecBCD- RecFOR-independent pathway of homologous recombination in Escherichia coli // 4th Congress of Croatian Geneticists with International Participation - Book of Abstracts / Šarčević H, Ugarković Đ, Vujaklija D et al. (ed.). Zagreb: Croatian Genetic Society, 2018. p. 25-25 (lecture)
Đermić E, Zahradka D, Vujaklija D, Đermić D. A new role of RecA protein in Escherichia coli? // 4th Congress of Croatian Geneticists with International Participation : Book of Abstracts / Šarčević H, Ugarković Đ, Vujaklija D et al. (ed.). Zagreb: Croatian Genetic Society, 2018. str. 44-44 (poster)
Repar J, Supek F, Klanjscek T, Warnecke T, Zahradka K, Zahradka D. Genome rearrangements in radiation-resistant bacteria // 4th Congress of Croatian Geneticists with International Participation : Book of Abstracts / Šarčević H, Ugarković Đ, Vujaklija D et al. (ed.). Zagreb: Croatian Genetic Society, 2018. str. 24-24 (lecture)
Đermić E, Zahradka D, Vujaklija D, Ivanković S, Đermić D. 3'-terminal Overhangs Regulate DNA Double-Strand Break Processing in Escherichia coli // EMBO Conference on Meiosis 2017 : Book of Abstracts / Tolić I (ed.). 2017. p. 93-93 (poster)
Hlevnjak A, Paradžik T, Kazazić S, Filić Ž, Zahradka D, Vujaklija D. Phosphorylation of the SSB protein from E. coli // John Innes/Rudjer Bošković Summer School in Applied Molecular Microbiology: Microbial Diversity and Specialised Metabolites. Dubrovnik, Croatia, 10.09.2016-18.09.2016 (poster)
Repar J. Elevated rate of genome rearrangements in radiation resistant bacteria // CEGSDM 2016 - Central European Genome Stability and Dynamics Meeting. Zagreb, Croatia, 15.10.2016-16.10.2016 (lecture)
Zahradka K, Hlevnjak A, Repar J, Buljubašić M, Zahradka D. Genetic differences in postsynaptic stages of two recombinational repair pathways in Escherichia coli // EEMGS 2016 Annual Meeting - Programme & Abstract book / EEMGS (ed.). Kopenhagen, 2016. str. 146-146 (poster)

